Surface options:
Viewer options:
More options:
Ligands
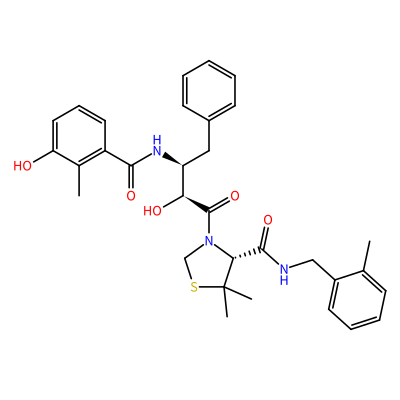
JE2_A_701
S1CN(C(=O)[C@@H](O)[C@@H](NC(=O)c2cccc(O)c2C)Cc3ccccc3)[C@H](C(=O)NCc4ccccc4C)C1(C)C
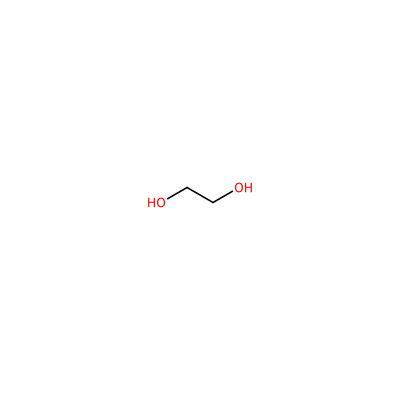
EDO_A_801
OCCO
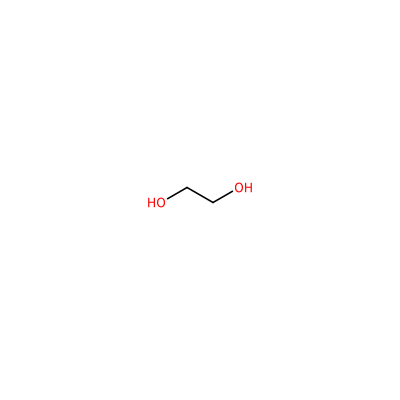
EDO_A_803
OCCO
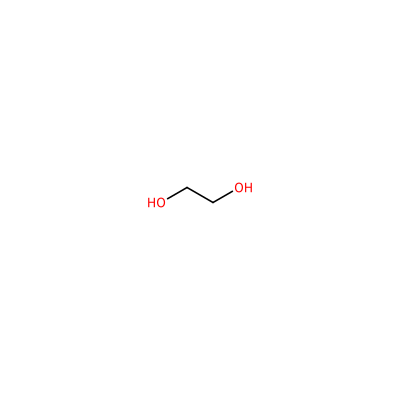
EDO_A_804
OCCO
EDO_A_805
OCCO
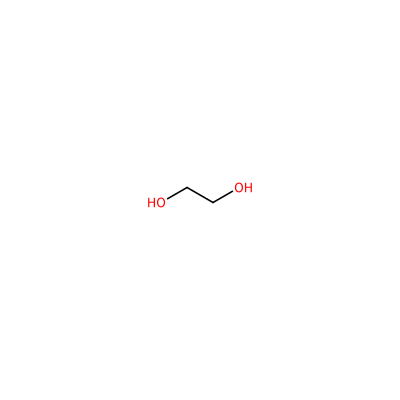
EDO_B_802
OCCO
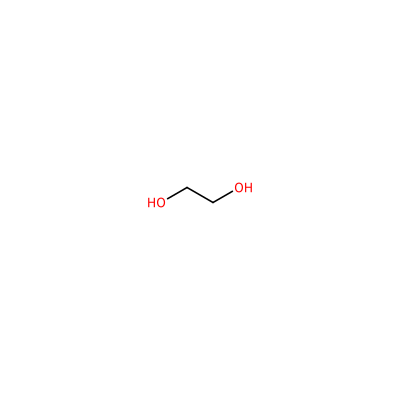
CL_A_902
[Cl-]
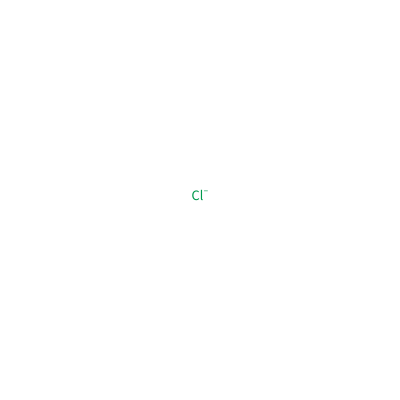
CL_A_905
[Cl-]
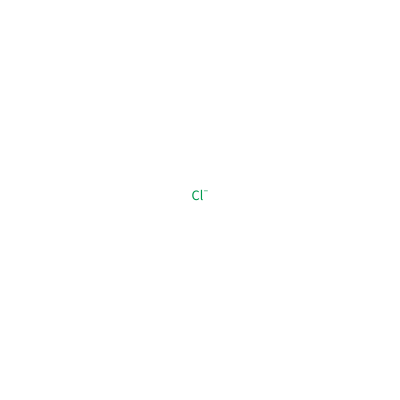
CL_B_901
[Cl-]
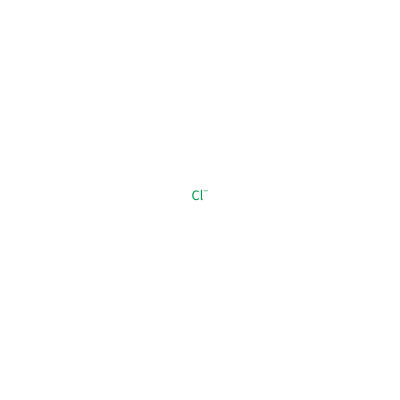
CL_B_903
[Cl-]
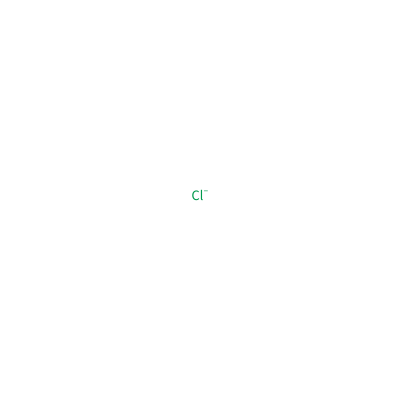
CL_B_904
[Cl-]
Pockets
calculating Pockets, this may take a minute
PoseEdit Interactive 2D ligand interaction diagrams
PoseEdit1 uses the PoseView algorithm2,3,4
and the InteractionDrawer JavaScript library (https://github.com/rareylab/InteractionDrawer) for the fully automatic drawing of highly interactive 2D ligand interaction diagrams.
Structures are depicted according to IUPAC recommendations.
Calculated interactions (hydrogen bonds, cation-pi interactions, pi-stackings, ionic interactions, and metal interactions)
of the ligand with nucleic acids, amino acids, and metals are depicted as colored dashed lines.
Hydrophobic contacts are visualized by labeled green splines.
The binding sites and the interactions occurring within were calculated by Protoss5, DoGSite36, and GeoMine7,8,9.
1. Diedrich, K.; Krause, B.; Berg, O.; Rarey, M., PoseEdit: enhanced ligand binding mode communication by interactive 2D diagrams. J Comput Aided Mol Des 2023, 37, 491-503. DOI:
https://doi.org/10.1007/s10822-023-00522-4
2. Stierand, K.; Maass, P. C.; Rarey, M.,
Drawing the PDB - Protein-Ligand Complexes in two Dimensions.
Medicinal Chemistry Letters 2010, 1 (9) 540-545. DOI:
https://doi.org/10.1021/ml100164p
3. Stierand, K.; Rarey, M.,
From Modeling to Medicinal Chemistry: Automatic Generation of Two-Dimensional
Complex Diagrams. ChemMedChem 2007, 2 (6), 853-860. DOI:
https://doi.org/10.1002/cmdc.200700010
4. Stierand, K.; Maass, P. C.; Rarey, M.,
Molecular complexes at a glance: automated generation of two-dimensional
complex diagrams. Bioinformatics 2006, 22 (14), 1710-6. DOI:
https://doi.org/10.1093/bioinformatics/btl150
5. Bietz, S.; Urbaczek, S.; Schulz, B.; Rarey, M.,
Protoss: a holistic approach to predict tautomers and protonation states
in protein-ligand complexes. J Cheminform 2014, 6, 12. DOI:
https://doi.org/10.1186/1758-2946-6-12
6. Graef, J.; Ehrt, C.; Rarey, M.,
Binding Site Detection Remastered: Enabling Fast, Robust, and Reliable Binding Site
Detection
and Descriptor Calculation with DoGSite3.
J Chem Inf Model 2023, 63 (10), 3128–3137. DOI:
https://doi.org/10.1021/acs.jcim.3c00336
7. Diedrich, K.; Ehrt, C.; Graef, J.; Poppinga, M.; Ritter, N.; Rarey, M.,
User-centric design of a 3D search interface for protein-ligand complexes.
J Comput Aided Mol Des 2024, 38 (23). DOI:
https://doi.org/10.1007/s10822-024-00563-3
8. Graef, J.; Ehrt, C.; Diedrich, K.; Poppinga, M.; Ritter, N.;
Rarey, M., Searching Geometric Patterns in Protein Binding Sites and
Their Application to Data Mining in Protein Kinase Structures.
J Med Chem 2022, 65 (2), 1384-1395. DOI:
https://doi.org/10.1021/acs.jmedchem.1c01046
9. Diedrich, K.; Graef, J.; Schoning-Stierand, K.; Rarey,
M., GeoMine: interactive pattern mining of protein-ligand interfaces
in the Protein Data Bank. Bioinformatics 2021, 37 (3), 424-425. DOI:
https://doi.org/10.1093/bioinformatics/btaa693
